CLiPS has had multiple successful projects in Clinical NLP, described below.
Accumulate
Accumulate (Acquiring Crucial Medical information Using Language Technology) is a large SBO-IWT funded program focusing on developing fundamental technology for the processing of clinical language in English and Dutch. The project is a cooperation with the University of Leuven and the university hospitals of Antwerp and Leuven. At CLiPS we have intensively worked on the following:
- spelling normalization,
- concept and relation extraction,
- negation and speculation detection,
- patient representation learning,
- question answering,
- interpretability,
- ethics and
- clinical prediction tasks.
Previous projects
The computational linguistics group of the CLiPS (CLiPS-CLG) research centre has a long history in biomedical Natural Language Processing, starting with the multidisciplinary EU FP6 project BIOMINT (Biological Text Mining, 2003-2006), where we developed Natural Language Processing (NLP) tools for knowledge extraction on medical text in cooperation with the Universities of Leuven, Geneva (SIB), Manchester, and Vienna. We continued this line of research with the BOF GOA project Biograph (Text Mining with Heterogeneous Knowledge Bases, 2007-2011), coordinated by CLiPS, where graph-based data mining techniques were applied to a combination of structured background knowledge (ontologies) and relations between concepts extracted from biomedical text (Medline). In that project, we successfully cooperated with the data mining and molecular genetics departments of the University of Antwerp and focused on deep analysis of biomedical text (e.g. looking at markers of uncertainty like negation and modality). We have also recently finished an IWT PhD project on clinical coding (Elyne Scheurwegs: Data fusion and structured input and output Machine Learning techniques for automated clinical coding, 2014-2017) in cooperation with the data mining research of the computer science department and the Antwerp University Hospital. CLiPS has also participated in a VLAIO Baekeland PhD project with the company Lynxcare as a partner.
In addition, our group has also been active in international benchmarking competitions on biomedical and clinical NLP, both from the side of their organisation (for example the CLEF 2012 and 2013 Labs on Question Answering for Machine Reading of biomedical texts about Alzheimer disease, Rome and Valencia) as from the side of participation, for example CEGS N-GRID shared task 2016 about psychiatric symptom severity identification and the 2018 n2c2 shared task on cohort selection.
People
Publications
Pieter Fivez, Simon Šuster and Walter Daelemans (2021) Scalable Few-Shot Learning of Robust Biomedical Name Representations. BioNLP, NAACL.
Pieter Fivez, Simon Šuster and Walter Daelemans (2021) Integrating Higher-Level Semantics into Robust Biomedical Name Representations. Workshop on Health Text Mining and Information Analysis (LOUHI), EACL.
Pieter Fivez, Simon Šuster and Walter Daelemans (2021) Conceptual Grounding Constraints for Truly Robust Biomedical Name Representations. EACL.
Madhumita Sushil, Simon Šuster and Walter Daelemans (2021) Are we there yet? Exploring clinical domain knowledge of BERT models. BioNLP Workshop, NAACL.
Madhumita Sushil, Simon Šuster and Walter Daelemans (2021) Contextual explanation rules for neural clinical classifiers. BioNLP Workshop, NAACL.
Madhumita Sushil, Simon Šuster and Walter Daelemans (2020) Distilling neural networks into skipgram-level decision lists arXiv preprint arXiv:2005.07111.
Simon Šuster, Madhumita Sushil and Walter Daelemans (2019) Why can’t memory networks read effectively? arXiv preprint arXiv:1910.07350.
Stéphan Tulkens, Simon Šuster and Walter Daelemans (2019) Unsupervised Concept Extraction from Clinical Text through Semantic Composition. Journal of Biomedical Informatics.
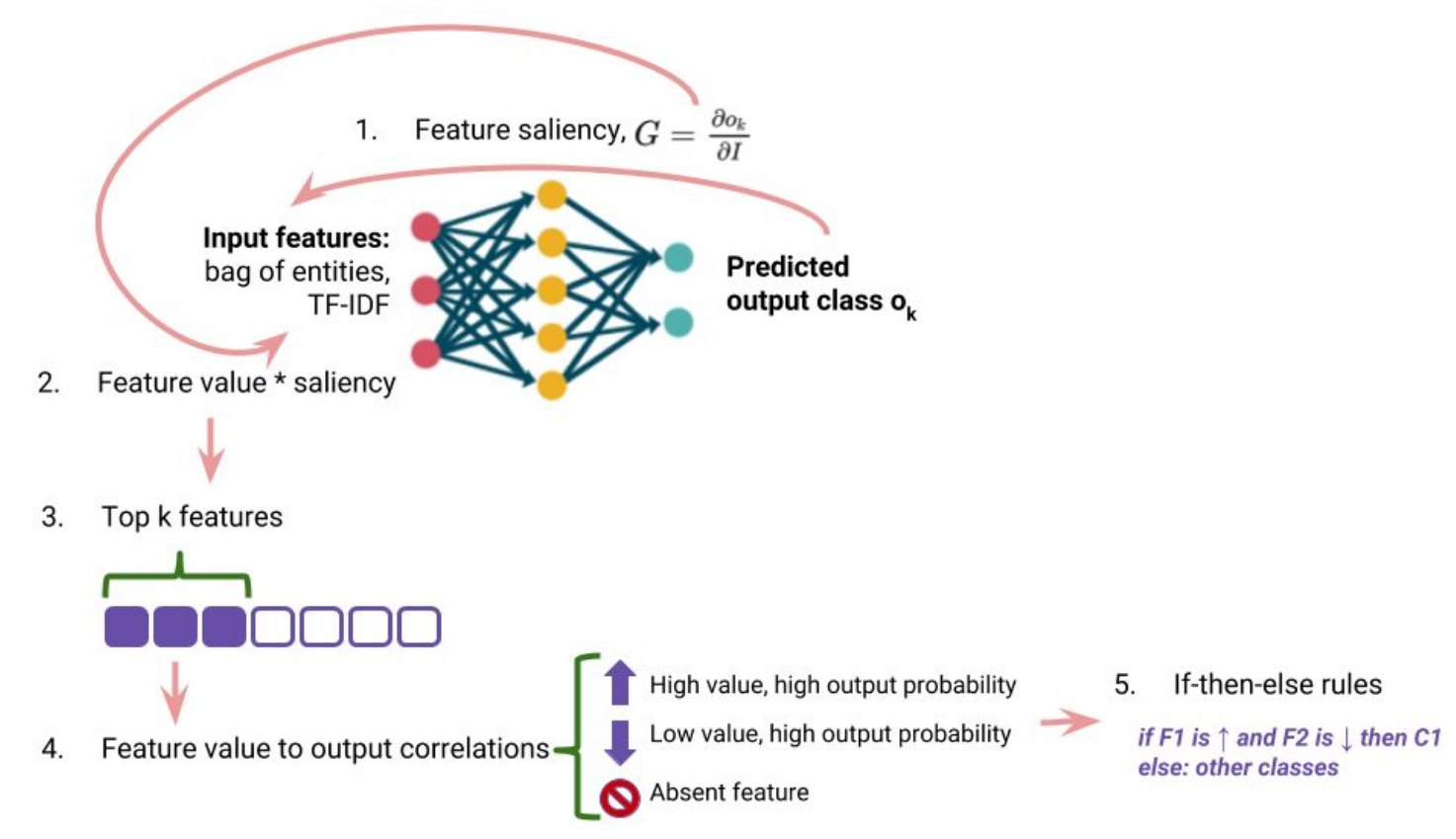
Madhumita Sushil, Simon Šuster and Walter Daelemans (2018) Rule induction for global explanation of trained models. Analyzing and interpreting neural networks for NLP (BlackBoxNLP), EMNLP.
Simon Šuster, Madhumita Sushil and Walter Daelemans (2018) Revisiting neural relation classification in clinical notes with external information. In Workshop on Health Text Mining and Information Analysis (LOUHI), EMNLP.
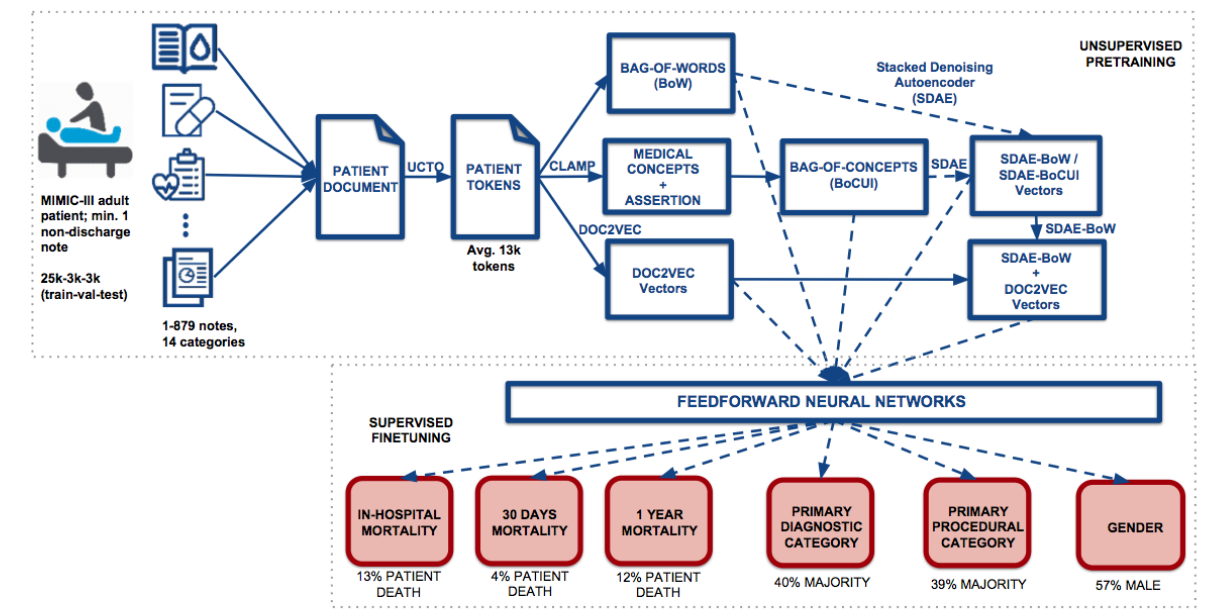
Madhumita Sushil, Simon Šuster, Kim Luyckx, and Walter Daelemans (2018) Patient representation learning and interpretable evaluation using clinical notes. Journal of Biomedical Informatics 84 (2018): 103-113.
Simon Šuster and Walter Daelemans (2018) CliCR: A Dataset of Clinical Case Reports for Machine Reading Comprehension. Proceedings of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies.
Madhumita Sushil, Simon Šuster, Kim Luyckx and Walter Daelemans (2017) Unsupervised Dense Patient Representations from Unstructured Clinical Data with Interpretable Classification Decisions. Workshop on Machine Learning for Health, NIPS.
Pieter Fivez, Simon Šuster and Walter Daelemans (2017) Unsupervised Context-Sensitive Spelling Correction of English and Dutch Clinical Free-Text with Word and Character N-Gram Embeddings. Computational Linguistics in the Netherlands Journal.
Elyne Scheurwegs, Madhumita Sushil, Stéphan Tulkens, Walter Daelemans, Kim Luyckx (2017) Counting trees in Random Forests: Predicting symptom severity in psychiatric intake reports. Journal of Biomedical Informatics.
Pieter Fivez, Simon Šuster and Walter Daelemans (2017) Unsupervised Context-Sensitive Spelling Correction of Clinical Free-Text with Word and Character N-Gram Embeddings. Workshop on Biomedical Natural Language Processing (BioNLP).
Simon Šuster, Stéphan Tulkens and Walter Daelemans (2017) A Short Review of Ethical Challenges in Clinical Natural Language Processing. First Workshop on Ethics in NLP.
Stéphan Tulkens, Simon Šuster and Walter Daelemans. (2016) Using Distributed Representations to Disambiguate Biomedical and Clinical Concepts. Workshop on Biomedical Natural Language Processing (BioNLP).
More soon…
Abstracts
Merijn Beeksma, Madhumita Sushil, Pieter Fivez, Simon Šuster, Florian Kunneman, Ghiath Ghanem, Walter Daelemans, Antal van den Bosch. (2018) Mining gold from EMRs: using clinical notes to automate patient screenings. N2C2 Shared Task on Challenges in Natural Language Processing for Clinical Data.
Software
This is a summary of openly available software with links to GitHub. For licensing, see the individual repositories. Please get in touch with us if you’d like to commercialize our software.
Robust representations of biomedical names
Text normalization
- clinspell: Clinical spelling correction with word and character n-gram embeddings
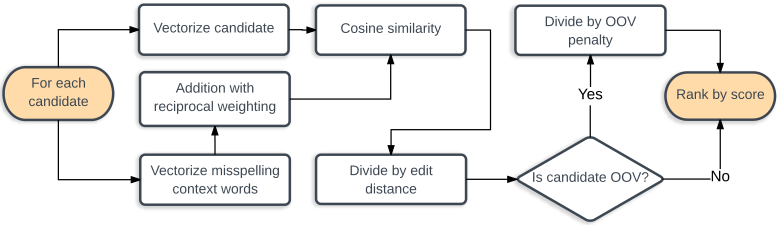
Concept extraction and disambiguation
- yarn: Disambiguating biomedical and clinical concepts with word embeddings
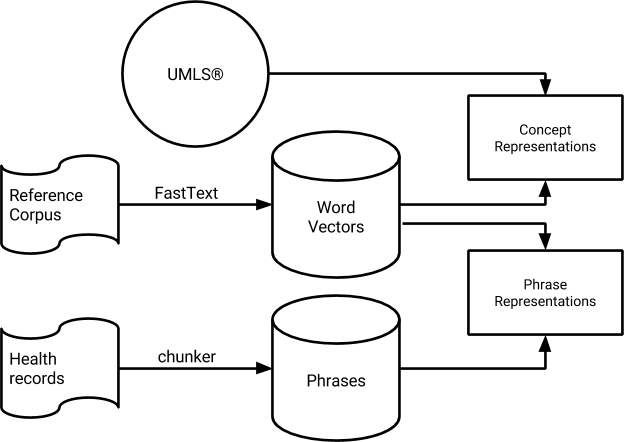
- conch: Extracting medical concepts and terminology from patient records, and linking them to UMLS CUI
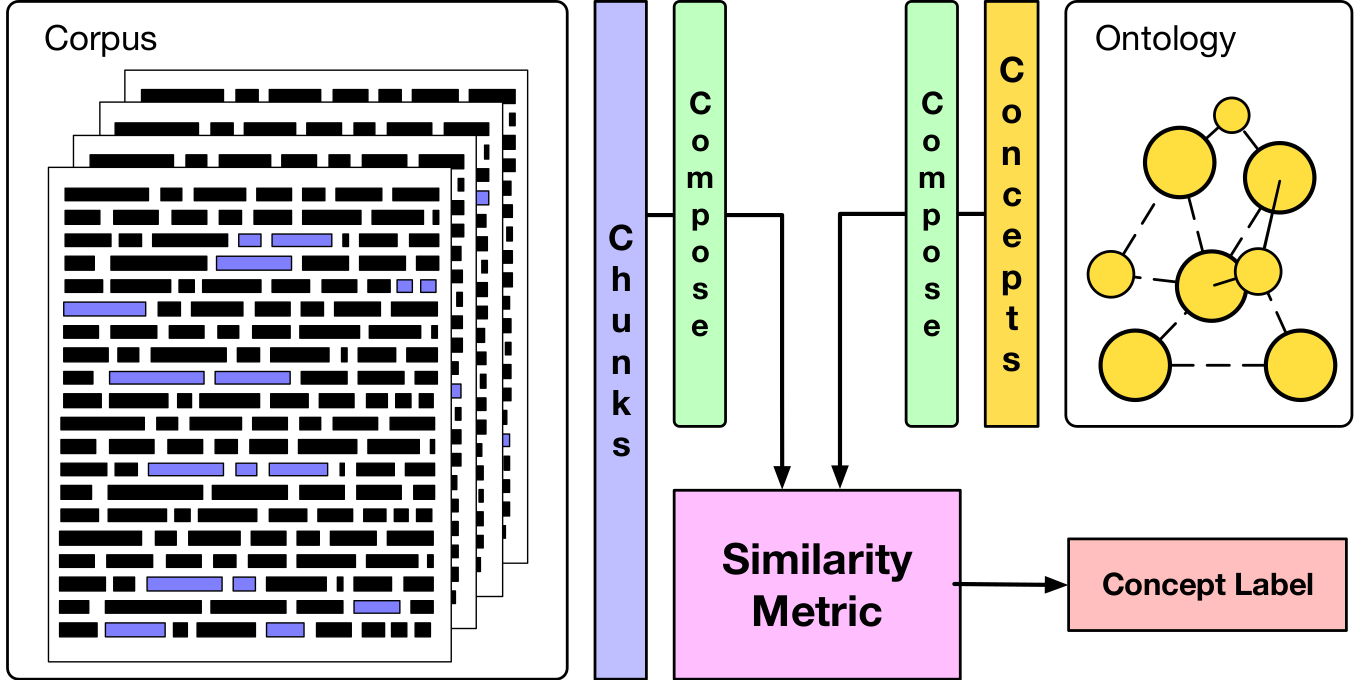
Negation detection
- nl-neg-clinical: Negation detection of concepts in Dutch clinical text
- en-neg-clinical: Negation detection of concepts in English clinical text
Speculation detection
- nl-spec-clinical: Speculation detection of concepts in Dutch clinical text
- en-spec-clinical: Speculation detection of concepts in English clinical text
Relation extraction
- clinical-rel-class: Revisiting neural relation classification in clinical notes with external information
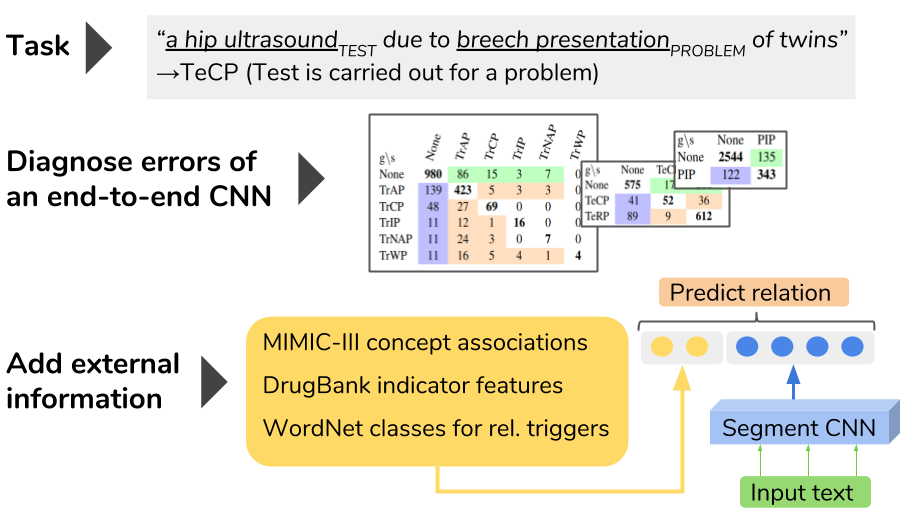
Patient representation learning
- pt-rep: Unsupervised patient representations from clinical notes with interpretable classification decisions
Interpretability
- interpret: Rule induction for global explanation of trained models
- Unravel Distilling neural networks into skipgram-level decision lists
Question answering and reading comprehension
- clicr: Machine reading and question answering on clinical case reports

- memory-networks: Memory networks for medical machine reading